ZMP
NP_001107902.1
Ensembl ID:
Description:
solute carrier family 2 (facilitated glucose transporter), member 11b [Source:RefSeq peptide;Acc:NP
Human Orthologues:
AP000350.10, SLC2A11, SLC2A5, SLC2A7, SLC2A9
Human Descriptions:
solute carrier family 2 (facilitated glucose transporter), member 11 [Source:HGNC Symbol;Acc:14239]
solute carrier family 2 (facilitated glucose transporter), member 7 [Source:HGNC Symbol;Acc:13445]
solute carrier family 2 (facilitated glucose transporter), member 9 [Source:HGNC Symbol;Acc:13446]
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 [Source:HGNC Symbol;Acc
solute carrier family 2 (facilitated glucose transporter), member 7 [Source:HGNC Symbol;Acc:13445]
solute carrier family 2 (facilitated glucose transporter), member 9 [Source:HGNC Symbol;Acc:13446]
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 [Source:HGNC Symbol;Acc
Mouse Orthologues:
Slc2a5, Slc2a7, Slc2a9
Mouse Descriptions:
solute carrier family 2 (facilitated glucose transporter), member 5 Gene [Source:MGI Symbol;Acc:MGI:
solute carrier family 2 (facilitated glucose transporter), member 7 Gene [Source:MGI Symbol;Acc:MGI:
solute carrier family 2 (facilitated glucose transporter), member 9 Gene [Source:MGI Symbol;Acc:MGI:
solute carrier family 2 (facilitated glucose transporter), member 7 Gene [Source:MGI Symbol;Acc:MGI:
solute carrier family 2 (facilitated glucose transporter), member 9 Gene [Source:MGI Symbol;Acc:MGI:
Alleles
There are 3 alleles of this gene:
| Allele Name | Consequence | Status | Availability |
|---|---|---|---|
| sa1577 | Nonsense | Available for shipment | Available now |
| sa13882 | Nonsense | Available for shipment | Available now |
| sa13746 | Nonsense | Available for shipment | Available now |
Mutation Details
Allele Name:
sa1577
Status:
Available for shipment
For more information about the meaning of this status and other statuses, please see our FAQs.
For more information about the meaning of this status and other statuses, please see our FAQs.
Availability:
Mutation:
C > A
Consequence:
Nonsense
Transcripts:
| Transcript ID | Consequence | Amino Acid Affected | Amino Acid Total | Exon Affected | Exon Total |
|---|---|---|---|---|---|
| ENSDART00000092333 | Nonsense | 66 | 504 | 3 | 12 |
| ENSDART00000132262 | None | None | 314 | None | 7 |
The following transcripts of ENSDARG00000063288 do not overlap with this mutation:
The following genes are also affected by this mutation:
Genomic Location (Zv9):
Chromosome 5 (position 10927739)
Other Location(s):
| Assembly | Chromosome | Position |
|---|---|---|
| GRCz10 | 5 | 9515499 |
| GRCz11 | 5 | 10020137 |
| GRCz12tu | 5 | 10736105 |
KASP Assay ID:
554-1520.1 (used for ordering genotyping assays)
KASP Sequence:
TTCTCTCAGGCTGTGCAGAACTTTATTAATCAGACCTGGACGGAGCGCTA[C/A]AGCACTGAAATCTCCAGTGATATGTTAACCCTCCTCTGGTCCATCATCGT
Long Flanking Sequence:
ATAATTATGAAATGTCTTTTTATACATTTTTATTTCGGCTTTACTCAATTCACACTGAAATGAAACCAAATGTAAAATATTTCTTTGACGATTAGCCAAAGTGACAGTTTATTACACTATTTATTGTGTTATTATTTCATTTTGGCTAACACATAAACTTTATACAGTTTTACGGTTTGTCTACACTCAAAAAATTACTGTTGCTGCTTGTTCAAACTACTTATTTAAAATGAGTTAAAACAACACAATTCTTAAGTTTTATGGGGGACATCTTAATTGTTTCATGCTCAATCCACTTAAATTCGTAAAACCAATTAAGTTAACTGAATCGATTTGTGTTGGGACAAAATGAAGATATTTTGTGGAACCCAGCATTTGTTTGTCAGTGTATGACGCTGTTTTTTTAGTGAGCCAGTGTTTATTCCTCTCTCATACTGTATGTGTGTTTGTTTCTCTCAGGCTGTGCAGAACTTTATTAATCAGACCTGGACGGAGCGCTA[C/A]AGCACTGAAATCTCCAGTGATATGTTAACCCTCCTCTGGTCCATCATCGTCTCCATCTTCACTATCGGTGGGCTTGTAGGAGCGTCTGTCGGAGGAACACTGGCTATTAGATTCGGAAGGTAAACTGAAGATCAAAATGCTGTAAAGGAAATCATTAAATGTGCAGGCATCAGTTATGAAGATATTATACACTGTTGTTTGTTCAAATGACTTATTTAAAATGAGCTGAAACAACTCTTGAGCTTTTTGGGAGAAATTAATTGTTTTATGGTAACTGTTAACTTATTTCCTTCATGTTGTCCCAACACTAATGATTGTGTGGAACAAAGGATTTGTTTTACAGTGCTGACCTGATTGTTTTTTCAGGAAAGGGACTCTACTGTTCAACAACACCTTTGCTCTGTTGGCTGCTTTGTTCATGGGTTTAAGTTATCCTGCAAGCACTTTTGAGCTTTTAGTCATTGGACGCTTCCTGACAGGAGTAAATGCAGGTAAATTCA
Associated Phenotype:
This allele has been associated with this phenotype by genetic linkage analysis and may not be causal.
See FAQs for more info.
See FAQs for more info.
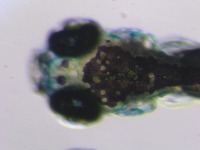
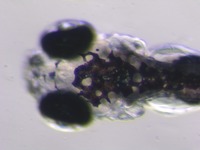
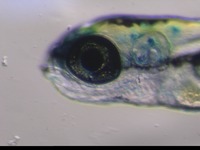
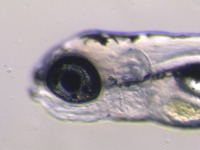


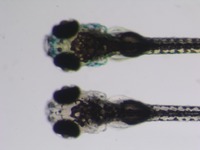
| Stage | Entity | Quality | Tag |
|---|---|---|---|
| Larval:Day 5 ZFS:0000037 |
iridophore ZFA:0009199 |
quality PATO:0000001 |
normal PATO:0000461 |
| Larval:Day 5 ZFS:0000037 |
melanocyte ZFA:0009091 |
quality PATO:0000001 |
normal PATO:0000461 |
| Larval:Day 5 ZFS:0000037 |
xanthophore ZFA:0009198 |
absent PATO:0000462 |
abnormal PATO:0000460 |
Mutation Details
Allele Name:
sa13882
Status:
Available for shipment
For more information about the meaning of this status and other statuses, please see our FAQs.
For more information about the meaning of this status and other statuses, please see our FAQs.
Availability:
Mutation:
T > A
Consequence:
Nonsense
Transcripts:
| Transcript ID | Consequence | Amino Acid Affected | Amino Acid Total | Exon Affected | Exon Total |
|---|---|---|---|---|---|
| ENSDART00000092333 | Nonsense | 128 | 504 | 4 | 12 |
| ENSDART00000132262 | None | None | 314 | None | 7 |
The following transcripts of ENSDARG00000063288 do not overlap with this mutation:
The following genes are also affected by this mutation:
Genomic Location (Zv9):
Chromosome 5 (position 10928172)
Other Location(s):
| Assembly | Chromosome | Position |
|---|---|---|
| GRCz10 | 5 | 9515932 |
| GRCz11 | 5 | 10020570 |
| GRCz12tu | 5 | 10736537 |
KASP Assay ID:
None (used for ordering genotyping assays)
KASP Sequence:
TTCAACAAMACCTTTGCTCTGTTGGCTGCTTTGTTCATGGGTTTRAGTTA[T/A]CCTGCAAGCACTTTTGAGCTTTTAGTCATTGGAYGCTTCMTGACAGGAGT
Long Flanking Sequence:
ACTGTATGTGTGTTTGTTTCTCTCAGGCTGTGCAGAACTTTATTAATCAGACCTGGACGGAGCGCTACAGCACTGAAATCTCCAGTGATATGTTAACCCTCCTCTGGTCCATCATCGTCTCCATCTTCACTATCGGTGGGCTTGTAGGAGCGTCTGTCGGAGGAACACTGGCTATTAGATTCGGAAGGTAAACTGAAGATCAAAATGCTGTAAAGGAAATCATTAAATGTGCAGGCATCAGTTATGAAGATATTATACACTGTTGTTTGTTCAAATGACTTATTTAAAATGAGCTGAAACAACTCTTGAGCTTTTTGGGAGAAATTAATTGTTTTATGGTAACTGTTAACTTATTTCCTTCATGTTGTCCCAACACTAATGATTGTGTGGAACAAAGGATTTGTTTTACAGTGCTGACCTGATTGTTTTTTCAGGAAAGGGACTCTACTGTTCAACAACACCTTTGCTCTGTTGGCTGCTTTGTTCATGGGTTTAAGTTA[T/A]CCTGCAAGCACTTTTGAGCTTTTAGTCATTGGACGCTTCCTGACAGGAGTAAATGCAGGTAAATTCAACACAAAATTTGAAGCACAACCATTTTAAACCACTTTGATGATGAGAAGAAATGTAAAAAATATGGACATTAGGTTGATTTCTGAAGGATGACTAGATTTCCATCCTAAAATGAAGTGACCTTTTTCAAAGTTTGCAAAAAAACATTTACATTTACATTTAGTCATTTAGCAGACGCTTTTATCCAAAGCGACTTACAAATGAGGACAAGGAAGCAATTTACACAACTATAAGAGCAACAATGAATAAGTGTTATAGGCAAGTTTCAGGTCTGTAAAGTCTAAGAAGGAAAGCATTAGTAATTTTTTTTATTTATTTTAAATGTTTTTGGTACAGTTAGTGTGATATCCAGAGAGGCAATTGCAGATTAGGAAGTGAAGTGGAGACTAAATAGTTGAGTTTTTAGTCGTTTCTTGAAAATAGCGAGTGACTCT
Associated Phenotype:
Not determined
Mutation Details
Allele Name:
sa13746
Status:
Available for shipment
For more information about the meaning of this status and other statuses, please see our FAQs.
For more information about the meaning of this status and other statuses, please see our FAQs.
Availability:
Mutation:
C > A
Consequence:
Nonsense
Transcripts:
| Transcript ID | Consequence | Amino Acid Affected | Amino Acid Total | Exon Affected | Exon Total |
|---|---|---|---|---|---|
| ENSDART00000092333 | Nonsense | 227 | 504 | 6 | 12 |
| ENSDART00000132262 | Nonsense | 37 | 314 | 1 | 7 |
The following transcripts of ENSDARG00000063288 do not overlap with this mutation:
Genomic Location (Zv9):
Chromosome 5 (position 10935154)
Other Location(s):
| Assembly | Chromosome | Position |
|---|---|---|
| GRCz10 | 5 | 9522914 |
| GRCz11 | 5 | 10027552 |
| GRCz12tu | 5 | 10743521 |
KASP Assay ID:
None (used for ordering genotyping assays)
KASP Sequence:
GCCGTCCTGCAGCTTCTGACKCTGCCCTGGTTCCCGGAGAGYCCTCGATA[C/A]CTGCTCATCGACCGAGGGGACGACATCGCCTGCCAGAAGGGTACAAACAC
Long Flanking Sequence:
AATGATCAACAAGAATTCAAGTTATTATTTATAAAGATATAAAAATAAAAATCACACTCGCAGAATTACTATATTATTTATTAATAAGGCACAGCCTTATTCTGGGTTCACACCTAAAGTGAATTAAGGTATTCCAGTAAATAACAAGTTGCAAAGTCACGTTTGCTGCGAATGTGTGGAATTCACCACAAGCAAGTCTTCTGCACAAGTTCAGCTTGATTGAATAATTTGTGAGGCAAAAACCATCCTGTAAGTAATGCTGTGTTCACACAAGACAAGACTGCTTCACTTGCTCTGGTTTACTCATGGGATGTTTTTCCCCTCATGCAATTTTTATGCATGTTTACAGTTGCATGGCAGTAATGTGTATTGTTGTTGTTTGTGTATGTCAGAGAATTGTTAGGAAAGGATGAGTACTGGCCGATCCTCTTGTCCACCATCTGTATTCCAGCCGTCCTGCAGCTTCTGACTCTGCCCTGGTTCCCGGAGAGTCCTCGATA[C/A]CTGCTCATCGACCGAGGGGACGACATCGCCTGCCAGAAGGGTACAAACACTTTCATAACTCTTCGCTTTGAGGATATTTTTATTTTATGATTAAAAACATAGTGGGCCCTATCATACACCCGGCGCAATGTGGCGCAAGGCACGGCGCAATAGTCTTTTGGTAGTTTCAGCTTGGCACAAGAGTCGTTTTGAGGCGTTGCGCTACGCTGTTTAATTAGCAAATGCATTAGTGCTCATTTGTGCGCCCATAGGCGTTCTGGTCTAAAATGAAGGCGTTCTGAGGCGCACTGCTGGCGCGTTGCTATTTTGAGAAACTATAATAGATTTTTCATTAGACCAAACAAACCCGGTCTAAACTCCGGCGCGGAGTTGCGCCTCACTTACACACTGCTTAATACGCACAAGAGAGCAATAGGCAAATATCTTTACATATGAAAAAATGTAAATATTAAGGATATATATATAGTATATAATAAGAATAGATATAGAATATAAATATA
Associated Phenotype:
Not determined
