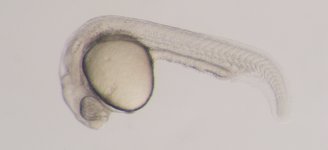
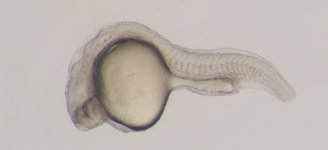
ZMP
lamc1
Ensembl ID:
ZFIN ID:
Description:
Laminin subunit gamma-1 [Source:UniProtKB/Swiss-Prot;Acc:Q1LVF0]
Human Orthologue:
LAMC1
Human Description:
laminin, gamma 1 (formerly LAMB2) [Source:HGNC Symbol;Acc:6492]
Mouse Orthologue:
Lamc1
Mouse Description:
laminin, gamma 1 Gene [Source:MGI Symbol;Acc:MGI:99914]
Alleles
There are 7 alleles of this gene:
| Allele Name | Consequence | Status | Availability |
|---|---|---|---|
| sa379 | Essential Splice Site | Available for shipment | Available now |
| sa19814 | Essential Splice Site | Available for shipment | Available now |
| sa9866 | Nonsense | Available for shipment | Available now |
Mutation Details
Allele Name:
sa379
Status:
Available for shipment
For more information about the meaning of this status and other statuses, please see our FAQs.
For more information about the meaning of this status and other statuses, please see our FAQs.
Availability:
Mutation:
G > A
Consequence:
Essential Splice Site
Transcripts:
| Transcript ID | Consequence | Amino Acid Affected | Amino Acid Total | Exon Affected | Exon Total |
|---|---|---|---|---|---|
| ENSDART00000004277 | Essential Splice Site | 325 | 1593 | 5 | 28 |
Genomic Location (Zv9):
Chromosome 2 (position 35675287)
Other Location(s):
| Assembly | Chromosome | Position |
|---|---|---|
| GRCz10 | 2 | 35971997 |
| GRCz11 | 2 | 35954454 |
| GRCz12tu | 2 | 38930810 |
KASP Assay ID:
554-0359.1 (used for ordering genotyping assays)
KASP Sequence:
AATATTTTTTATAGGCACATTTTCTGACTGGTGTTTGTGTGTGTGTTTCA[G/A]CTTGTAACTGTAATGGAAAGAGCGCTGAATGTTACTTCGACCCTGAGCTG
Long Flanking Sequence:
TCTGATTGTTTGACATTAGAAAAATAAACATGTATAGCTAAGAGAACCTGAAAAGTTTTTCAAAATATGGAATCCCTATATAGGGTTTCTGAAGGAAGCTGAAATGAATCTTCTGTCCATATAACTTGTATCTTGTATCACCTGCTATTAATTTATCATTGTTTTGATACATTTATTTTGTATGTGTATGTACATGCATTTATTTATTTATATACTTGTAATGGAAATTTTTATTCTTTTTTTTATTTATTTATTATTATTTTTTTTTTTACTTTTTTTTAATATTCTTAACTGCGACCCTCTTACCGTTAGTTTACTATGAGGGACTGATGCACTGAAACAAAGCCCTGCCCCCTGTTAAATATTTAGTTTCAGTTGAAACTACATACAAAATACTGAAATAAAGTCTCAGTAACTTCCAGTTCACTTTAAACCTTCACTTTTCAGACTAATATTTTTTATAGGCACATTTTCTGACTGGTGTTTGTGTGTGTGTTTCA[G/A]CTTGTAACTGTAATGGAAAGAGCGCTGAATGTTACTTCGACCCTGAGCTGTATCGTGCCACTGGTCACGGAGGTCACTGCCGTAACTGTGCTGACAATACAGATGGGCCGAAGTGTGAGCGATGCCTGGCCAACTACTACAGAGAGGCCTCCGGTCAGCGCTGTCTGTCCTGCGGCTGCAACCCTGTTGGTGAGTAACAGCAATTCAGAAGGCTCAGGGTGGAAGATGCATCAATAGCCAAGCATATTATATTTTGCTTTGCTTTACTTTTTCCCCTTGGTTTTGTTGTTCATTTGTTTCTTTGTACATTCATGTAGTTTTGCTAGGTGTTTTTTTTTTTTTTGTCGTCTTTGTTTTTGTTTTCTCAACTTTTTTTTTCTTTTGTTTTTGGTTCTCAGTTGTTTCTTTGTACATGCGTGTTTGACGGTTTTGTTTTGTTTTTGTCTTCATTTGTTTTGTTTTCTTTTTTTCTCAACTTTTTTATCTTTTGTTTTTGTTAT
Associated Phenotype:
This allele has been associated with this phenotype by genetic linkage analysis and may not be causal.
See FAQs for more info.
See FAQs for more info.
| Stage | Entity | Quality | Tag |
|---|---|---|---|
| Pharyngula:Prim-5 ZFS:0000029 |
brain ZFA:0000008 |
quality PATO:0000001 |
abnormal PATO:0000460 |
| Pharyngula:Prim-5 ZFS:0000029 |
extension ZFA:0000106 |
quality PATO:0000001 |
abnormal PATO:0000460 |
| Pharyngula:Prim-5 ZFS:0000029 |
eye ZFA:0000107 |
quality PATO:0000001 |
abnormal PATO:0000460 |
| Pharyngula:Prim-5 ZFS:0000029 |
notochord development GO:0030903 |
disrupted PATO:0001507 |
abnormal PATO:0000460 |
| Pharyngula:Prim-5 ZFS:0000029 |
somite ZFA:0000155 |
quality PATO:0000001 |
abnormal PATO:0000460 |
Mutation Details
Allele Name:
sa19814
Status:
Available for shipment
For more information about the meaning of this status and other statuses, please see our FAQs.
For more information about the meaning of this status and other statuses, please see our FAQs.
Availability:
Mutation:
A > T
Consequence:
Essential Splice Site
Transcripts:
| Transcript ID | Consequence | Amino Acid Affected | Amino Acid Total | Exon Affected | Exon Total |
|---|---|---|---|---|---|
| ENSDART00000004277 | Essential Splice Site | 427 | 1593 | 7 | 28 |
Genomic Location (Zv9):
Chromosome 2 (position 35676756)
Other Location(s):
| Assembly | Chromosome | Position |
|---|---|---|
| GRCz10 | 2 | 35973466 |
| GRCz11 | 2 | 35955923 |
| GRCz12tu | 2 | 38932279 |
KASP Assay ID:
None (used for ordering genotyping assays)
KASP Sequence:
None
Flanking Sequence:
CGGACACTTCTGAAATTGCTGTGTCTAAAGATTCCTCTTTTTCTCTTCAC[A/T]GACCGTGCTCTTGTAATCCAGCCGGAAGCACACAGGAGTGTGATGTCCAG
Long Flanking Sequence:
GTAAAAAAGTTTCACAAAACAAATCCAATTTATAATATAATTTTTGTGATTTGTTTATTTGTTTGTTTGTTTGTAAAATACATTAGAGGTAAGATAATTACAGATAGATTGTCAGACACCTGAAACAATAGGGGTAAAGTTTTCCTTTGCTTTTAATTTGAAGTAAAGAACTTCATTTGGTACCATCATTTTCTATGAATTGGTTATAAATACAAAATATATTACCTGAAAATGGCTTTGTTTTGTAACCATTTGTCTGATTTGCATCTGCAGGGTCTCTCAGCACTCAGTGTGATAACACTGGCAGATGCAGCTGTAAGCCAGGAGTGATGGGAGACAAGTGTGACAGATGCCAACCTGGATACCACTCGCTCTCTGAGGCAGGATGCAGGTAAAGATGTCTTTAACAACCAACCTGGAACACTTTGAACTTTTAAAGCACCTCAGATGCGGACACTTCTGAAATTGCTGTGTCTAAAGATTCCTCTTTTTCTCTTCAC[A/T]GACCGTGCTCTTGTAATCCAGCCGGAAGCACACAGGAGTGTGATGTCCAGACCGGACGCTGCCAGTGCAAGGAAAATGTGGACGGATTCAACTGTGACCGGTCAGACTCATTACTAGAGCAAATATCCGTGCACATGTTCACACACAGTAGCAACTGTTGGGGCTTTTTAAAGTCCCCTGTGCTGTTAGCGATGTCCACACTTGACATGTTCACAAGGACAAAAGCGTATAATTGCTTGATTTGTTTTAAAAATGTTCTCAAATTAGAAGCAAATGGTCAGCAGAATTAACCTGAACATCAACACTCCAGCAGCCCCATAAAGACCGTCCGGCTTTCAGCTCCTGCTCTGTTATGAGGACGTCTGCTGGAGCTTTAGAAACAGACATTTATAAGGGGCTTCTGACGTCTACACTGTTTTTCTTTCGGAAAGTCCCTGTGTGGGTGACACACGGAGGATCTGAGGCCCAGCCTGTGTGTGTGTTTTGTGTGTGTGTCTGAG
Associated Phenotype:
Not determined
Mutation Details
Allele Name:
sa9866
Status:
Available for shipment
For more information about the meaning of this status and other statuses, please see our FAQs.
For more information about the meaning of this status and other statuses, please see our FAQs.
Availability:
Mutation:
T > A
Consequence:
Nonsense
Transcripts:
| Transcript ID | Consequence | Amino Acid Affected | Amino Acid Total | Exon Affected | Exon Total |
|---|---|---|---|---|---|
| ENSDART00000004277 | Nonsense | 969 | 1593 | 17 | 28 |
Genomic Location (Zv9):
Chromosome 2 (position 35692130)
Other Location(s):
| Assembly | Chromosome | Position |
|---|---|---|
| GRCz10 | 2 | 35988840 |
| GRCz11 | 2 | 35971297 |
| GRCz12tu | 2 | 38947651 |
KASP Assay ID:
None (used for ordering genotyping assays)
KASP Sequence:
ATTTGGTTTTCCTGCGTTGACGCTTGGAATGTTTGTACAGCATGTGACTG[T/A]GACCCAGAAGGCTCAGAATCAGCTCAGTGTAARGAGGACGGCCGCTGTCA
Long Flanking Sequence:
CAACTTCTTCGGATTCAGTTCATCGGGTTGCAAACGTACGGTGCCTTATCATTGGCTTACTGTAGTCTCAAATTTCACAACCAAACTTTAGGTCTTAAATTCACTGAAGTATTGTTTATAGGTCCCAGAGAATGTGTGTTATGTGTTATTTTGTTTTGTCTAGTCTTGATTGTGGTGTTTGTTTGTTTTTCCACATATAATTCACTTTATTAGGACTACAAATGTGATCATCATCGTGATGTAGGCCTTAAGTTCAATTCTCAAAGTCTTAGATTTGACTTTGTAATACAGAAATCCTTGTCTAGCAATTGTGCTAGATGCAAAATTAATCATTAAAAGTCATAAATTACAAAAATTATATTTTAGGTGTTAAACATTTATTTTGGATTTCACTGTATTAAGACTACAAATAAATCCAACAGTCATGATATAGGTCTTTGTGCTATATGCATTTGGTTTTCCTGCGTTGACGCTTGGAATGTTTGTACAGCATGTGACTG[T/A]GACCCAGAAGGCTCAGAATCAGCTCAGTGTAAGGAGGACGGCCGCTGTCATTGTCGGCCAGGTTTTGTGGGCTCTCGCTGTGATATGTGTGAGGAAAACTACTTCTACAACCGCTCGACACCTGGCTGCCAGCAGTGCCCTAACTGTTACAGCCTGGTCAGAGACAAGGTGAGCCATTTTCTCATGAAACTAGAATGATTCGTTTTAGAGCATGAAGCAACTTCTAATACACCTGTTGCATTCCTGTACAGGTAAACCAGCAGAGACAAAAACTGCTTGACCTGCAGAATTTGATTGATAGTCTGGACAACACTGAGACGACAGTGAGCGACAAGGCCTTTGAGGACAGACTGAAGGAGGCAGAGAAAACTATAATGGACTTGCTGGAAGAGGCTCAGGCTAGCAAAGGTATAGATTCTTGTTATAGATTACAACAGCTCACTGTGTGATGAAGCCATTGAATGTAGTTATATGTCCAAAAAATTCAAGATATTGAGAAA
Associated Phenotype:
Not determined