ZMP
cyp26b1
Ensembl ID:
ZFIN ID:
Description:
Cytochrome P450 26B1 [Source:UniProtKB/Swiss-Prot;Acc:Q6EIG3]
Human Orthologue:
CYP26B1
Human Description:
cytochrome P450, family 26, subfamily B, polypeptide 1 [Source:HGNC Symbol;Acc:20581]
Mouse Orthologue:
Cyp26b1
Mouse Description:
cytochrome P450, family 26, subfamily b, polypeptide 1 Gene [Source:MGI Symbol;Acc:MGI:2176159]
Alleles
There is 1 allele of this gene:
| Allele Name | Consequence | Status | Availability |
|---|---|---|---|
| sa2 | Nonsense | Available for shipment | Available now |
Mutation Details
Allele Name:
sa2
Status:
Available for shipment
For more information about the meaning of this status and other statuses, please see our FAQs.
For more information about the meaning of this status and other statuses, please see our FAQs.
Availability:
Mutation:
A > T
Consequence:
Nonsense
Transcripts:
| Transcript ID | Consequence | Amino Acid Affected | Amino Acid Total | Exon Affected | Exon Total |
|---|---|---|---|---|---|
| ENSDART00000110347 | Nonsense | 46 | 511 | 1 | 6 |
Genomic Location (Zv9):
Chromosome 7 (position 26491727)
Other Location(s):
| Assembly | Chromosome | Position |
|---|---|---|
| GRCz10 | 7 | 25053473 |
| GRCz11 | 7 | 25324630 |
| GRCz12tu | 7 | 25313064 |
KASP Assay ID:
554-0011.1 (used for ordering genotyping assays)
KASP Sequence:
None
Flanking Sequence:
CCCAGCAGCTCTGGCAGCTGAGGTGGACCGCAACACGGGACAAGAGCTGC[A/T]AGCTGCCCATGCCCAAGGGCTCCATGGGGTTCCCCATCATCGGAGAAACA
Long Flanking Sequence:
GAAATTTAAAACAACGAATCAATCTCAAGTTGGACAAGTTCACTCCAAATTTACACTTCAAAAATTTGAATTTAATTGATGAATTCAAGAAATATTGGTCACCGATTTGGATGATTTAAAATGATATACACACATTTAAGATCGATTTGTTATATCCAGCATTCTGTTAATTAAATAGCCGACTATATTTTATTTATTCATTTACTCTACATAAACTTTTAGGGTTGGATTTGTGCTTTTTCGAATAATTGTCGCAGAGAGTCATATCTAACAGAGTAGGTCACTCTCCTAATTTTAGGTTTAACCACTTTATTGCTCATCACTCCAAAGAGATATTTGAGACAAGTCCCCGGACGTTCACAGCCATGCTCTTCGAGAGTTTTGACCTTGTCTCGGCGTTGGCGACGCTGGCTGCGTGTTTAGTGTCCATGGCACTTCTTCTGGCCGTGTCCCAGCAGCTCTGGCAGCTGAGGTGGACCGCAACACGGGACAAGAGCTGC[A/T]AGCTGCCCATGCCCAAGGGCTCCATGGGGTTCCCCATCATCGGAGAAACATGCCACTGGTTCTTTCAGGTAAGCGCTCATTCTCCTTGTGCCTTTCCAAGTTTCCAGTCTCCCTGCTGTCGCACGCACAGCGCTCCGGCGCGCCAGACACCGCGGGCTCGGCGCATATAGTTTTTCCTGCAGGTAACTGGGTTGTTAAAGACACTTCAAGAGTAGGATAACGTCCACTAATGCTTTTCGCACGTGTGCACTCGTGAGCAATATTGCGCTTTAAAATAAGAACAGCTTGCAGGTTAGTTATTCCCACGTGTGTAATTTCTTCCGTGCCATATTTCCATGGTGTTAATAAGCTGTCTGTTTTAGATAGCCGTCAGTTTTGTTTAATACAGCCAGTTCTCAATCCACGTGGAACAATCTGTTTGCGGCTGCCAAGGAAGGCGCACTCCGTTCCAGAAATATTGTAGTCCTTGGCTTCCGATGCGCGCGAGAACACAGCCACTG
Associated Phenotype:
This allele has been associated with this phenotype by genetic linkage analysis and may not be causal.
See FAQs for more info.
See FAQs for more info.
| Stage | Entity | Quality | Tag |
|---|---|---|---|
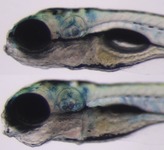
| Larval:Day 5 ZFS:0000037 |
swim bladder ZFA:0000076 |
aplastic PATO:0001483 |
abnormal PATO:0000460 |
| Larval:Day 5 ZFS:0000037 |
ventral mandibular arch ZFA:0001273 |
quality PATO:0000001 |
abnormal PATO:0000460 |